Study explores new tool for genome editing
By Krishna Ramanujan
Gene editing, a burgeoning technology that allows scientists to manipulate a specific region of DNA, has profound implications for medicine and agriculture. The technology makes use of a bacterial immune system known as CRISPR, an acronym for “clustered regularly-interspaced short palindromic repeats.”
Scientists believe there may be as many as five types of such CRISPR systems. So far, scientists have explored applications with a CRISPR/Cas9 system, which belongs to Type II.
A new Cornell study, published Feb. 10 in Nature, breaks down key mechanisms in a CRISPR Type I system, a significant step toward one day using this system for even more specific and accurate gene editing.
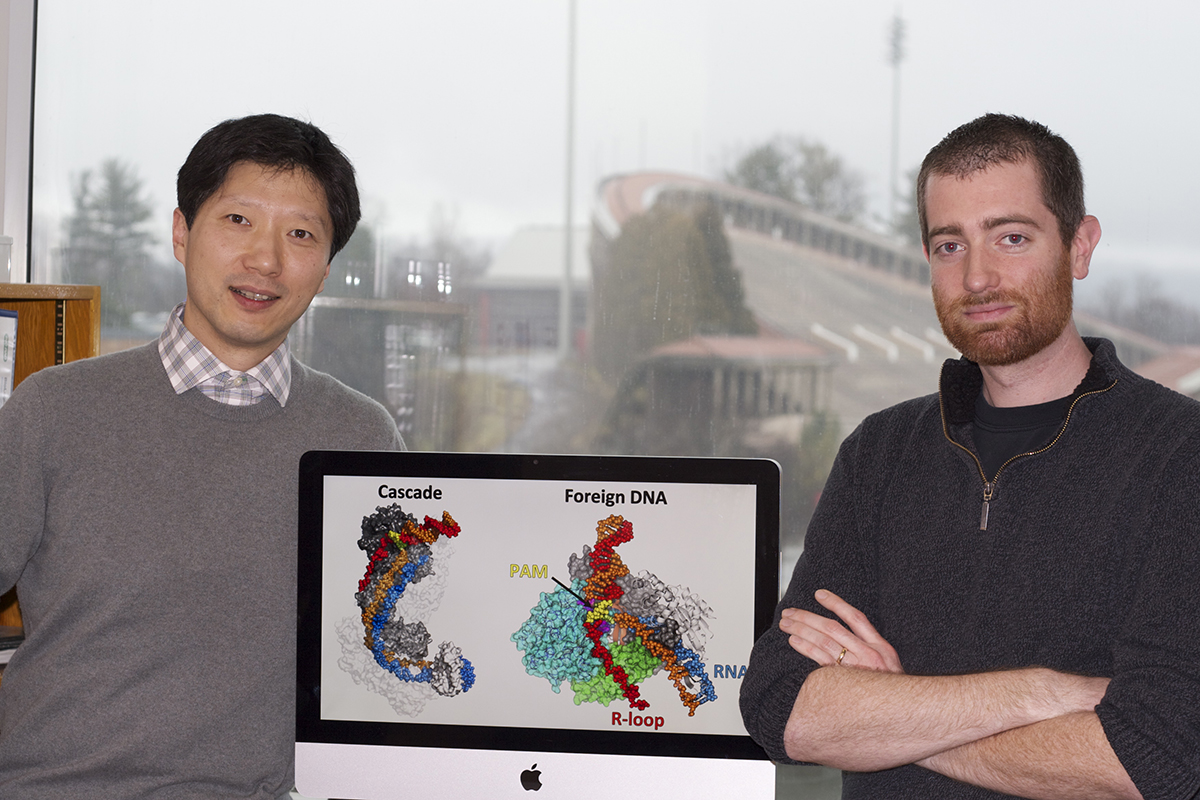
“Everyone is talking about Type II, but this is only the beginning, I think,” said Ailong Ke, associate professor of molecular biology and genetics in the College of Arts and Sciences, and the paper’s corresponding senior author. Robert Hayes, a postdoctoral fellow in Ke’s lab, is the paper’s first author.
“CRISPR systems are going to lead to a lot of big impacts, not only in the health industry but also in the agriculture industry, and that’s why we want to fully understand each of these systems and try to make the best use of them,” Ke added. Technology based on the CRISPR/Cas9 Type II system has been used to correct genetic disorders such as muscular dystrophy in mice, among other diseases.
All of the CRISPR systems utilize CRISPR RNA as a guide, though the specifics of how each system locates its targets vary greatly. RNA, the molecule most often used to transmit genetic information from DNA to proteins, has been modified to serve as a guide to direct CRISPR-associated proteins (Cas proteins) to a precise string of DNA. Once located, these proteins cut the viral DNA and disable the virus.
While the Cas9 Type II system targets a 20-nucleotide long DNA sequence, the Type I CRISPR seeks out a 32 to 35-nucleotide-long target, which will enable scientists to edit DNA sequences with greater specificity, while also reducing the risk of unintentional off-target effects.
This study used X-ray crystallography to determine the structure of a CRISPR Type I complex, called Cascade, that differentiates between “self” and “foreign” DNA, so that the CRISPR immune system can correctly attack viral DNA, while avoiding the bacteria’s own CRISPR sequence.
When a virus re-infects a bacterial cell, RNA guides Cascade to the viral DNA based on its memory of previous viral infections. A short DNA sequence that flanks the viral target, called PAM (protospacer adjacent motif), acts like a lock where Cascade is the key and allows Cascade to access the foreign viral DNA. Cascade then employs a cleaving enzyme called a nuclease to cut apart the viral DNA.
Researchers want to take advantage of these prokaryotic immune system mechanisms to locate specific DNA sequences found in diseases, cut out a specific nucleotide that causes that disorder, and replace it with a healthy version.
DNA molecules are made up of two strands of DNA entwined in a double helix. Each strand’s backbone lie closer together on one side of the helix than the other, creating a major groove where the backbones are far apart and a minor groove where they are close together. The grooves twist around the molecule on opposite sides.
The Cas9 enzyme in the Type II CRISPR system recognizes the PAM marker from the DNA major groove side and is very specific. But, in Type I systems, the Cascade molecule can recognize as many as five PAMs.
“With a structure snapshot of the Cascade recognizing the foreign DNA, we rationalized all the different PAMs that Cascade can recognize,” said Ke. “To everyone’s surprise, Cascade recognizes PAM from the DNA minor groove side. The recognition is inherently promiscuous. The research offers a first step toward engineering different PAM specificities, which will enable us to exploit Type I CRISPR system for a wider range of targets,” Ke said.
Co-authors include researchers from Johns Hopkins University and Montana State University.
The study was funded by the National Institutes of Health.
Media Contact
Get Cornell news delivered right to your inbox.
Subscribe