Cornell nano-researchers create component for a 'lab on a chip' that cuts DNA separation from a day to a matter of minutes
By Bill Steele
Researchers have long sought to create a "laboratory on a chip" that could greatly speed up the process of DNA sequencing. That goal has come a step closer with the announcement that Cornell University researchers have built and tested a nanofabricated device that can separate DNA fragments by length.
The silicon-based device performs the same function as the cumbersome gel electrophoresis process biologists now use but in as little as 15 to 30 minutes, rather than the 12 to 24 hours the gel process typically requires. It also can be more precisely controlled, the researchers say.
Harold Craighead, director of the Cornell Nanobiotechnology Center and professor of applied and engineering physics, and graduate student Jongyoon Han built on their previous research on the behavior of DNA in microscopic passageways to make the device, which fits on a silicon chip 15 millimeters (about three-quarters of an inch) long. They describe their work in the latest issue (May 11) of the journal Science.
A nanofabricated device like the one described in the paper could be incorporated into a complete analysis system on a single chip, the researchers say. In addition, they believe, it could be modified to separate various proteins and other molecules, such as inorganic polymers, in addition to DNA.
Separating DNA fragments by length is a fundamental part of the process of DNA sequencing to read out the genetic code of a sample, and is used in DNA "fingerprinting" to identify a person from a tissue sample or determine whether or not one person is related to another.
The traditional method is to place a sample at one end of a column of an organic gel and apply an electric field to the column, propelling the DNA fragments through the gel. As they slowly snake their way through the tiny pores of the material, fragments of different lengths move at different speeds and eventually collect in a series of bands as a ladderlike structure that can be photographed using fluorescent or radioactive tags.
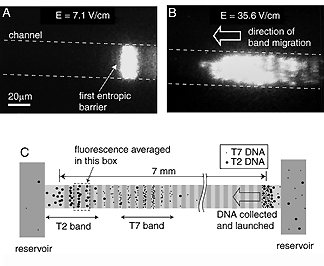
In previous work, Craighead and Han used the techniques developed for the manufacture of microscopic electronic circuit chips to carve on silicon tiny passageways so small that DNA strands could move through them only with difficulty. They built a channel consisting of alternating deep and shallow areas, placed DNA samples in water solution at one end of the channel and applied an electric field to draw the sample toward the other end. They saw that in the deep sections a chain-like DNA fragment contracts into a roughly spherical shape; but in this form it is too large to fit through the shallow sections. At the barrier between a deep and shallow section, the fragment is delayed until it can stretch out thin enough to fit through the smaller space.
Paradoxically, the researchers found that longer strands move through the device faster than shorter ones, the opposite of what happens in organic gels. The reason, they said, is that the longer strand, in the form of a larger sphere, presses a larger area against the barrier, meaning that there are more parts of itself that might extend into the shallow area to form a sort of beachhead and pull the rest through.
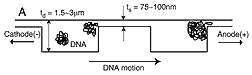
Armed with this knowledge, Han used the facilities of the Cornell Nanofabrication Facility to build a silicon device with channels made up of about 1,400 sections, each about 1.5 millionths of a meter deep, separated by shallow barrier sections only 75 to 100 billionths of a meter deep. When a DNA sample migrates through this device, fragments of different lengths travel at different speeds and arrive at the end in separate bands. Because the larger fragments move faster, it's like a marathon race in which a bunch of the fattest runners show up first, followed by successive groups of progressively thinner runners, with everyone of a particular size arriving more or less together. The DNA strands are tagged with a fluorescent material, and the peaks of fluorescence at the end of the channel measured over time form the readout.
In tests, the researchers ran standard reference samples of DNA, known as "ladders," through the device, and obtained results similar to running the same samples through a conventional organic gel.
Resolution of the nanofabricated device could be improved by making it longer, with more pockets, Han says. He also notes that this "artificial gel" allows more precise control in several ways than an organic gel: The depth of the channels can be precisely controlled, whereas in an organic gel the pore size will vary from place to place. The length of the deep sections of the channel also can be controlled, which determines whether or not a DNA strand has time to completely collapse back into a spherical shape before reaching the next barrier.
The device also has advantages for the separation of very long strands of DNA, the researchers said in their paper. Organic gels have a problem doing this, Han explains, because the strands must stretch out into a long, snakelike form to get through the pores of the gel, but then they never collapse back to spherical form; as a result, he says, strands of varying length all move through the gel at about the same speed. In the silicon device even very long strands can collapse to a spherical form in each deep section.
Finally, the new device makes it easy to extract separated DNA for further analysis: As a band of strands of a particular length arrives at the end of the channel, it can just be drawn off. Extracting samples from an organic gel requires complex chemical separation.
Related World Wide Web sites: The following sites provide additional information on this news release. Some might not be part of the Cornell University community, and Cornell has no control over their content or availability.
Press release about previous research on DNA trapping
http://www.news.cornell.edu/releases/Oct99/DNAtrapping.ws.html
Media Contact
Get Cornell news delivered right to your inbox.
Subscribe